News and Updates
| New publication: "Prosit" | 05/27/2019 | |
Prosit: Proteome-wide prediction of peptide tandem mass spectra by deep learning We are very excited to present the latest progress in the ProteomeTools project termed "Prosit". Prosit (https://www.proteomicsdb.org/prosit/) is a deep learning framework that offers free and easy generation of custom spectral libraries using very high quality predicted HCD MS2 spectra for any organism and protease as well as iRT prediction. Prosit is part of the ProteomeTools project and was trained on the project's high quality synthetic datasets.
| ||
| ProteomeTools @ ASMS 2019 Conference | 05/02/2019 |
| Annual meeting of the American Society for Mass Spectrometry The ProteomeTools team will be present at the ASMS 2019 Conference in Atlanta (GA, USA), June 2nd to 6th. We are looking forward to presenting new developments and to discussing the project. Join us for the following presentations: Oral:
Poster:
Usermeetings and Workshops:
For abstracts and additional information, please refer to the ASMS 2019 Final Program. | |
| ProteomeTools content presented on ASMS 2018 | 07/09/2018 |
Reprints of ProteomeTools content: We are happy to provide access to posters presented on ASMS 2018 in San Diego:
In case you have been attending ASMS, you can watch the webcast of the two oral contributions on the ASMS webcast website:
| |
| New Research Article | 05/30/2018 |
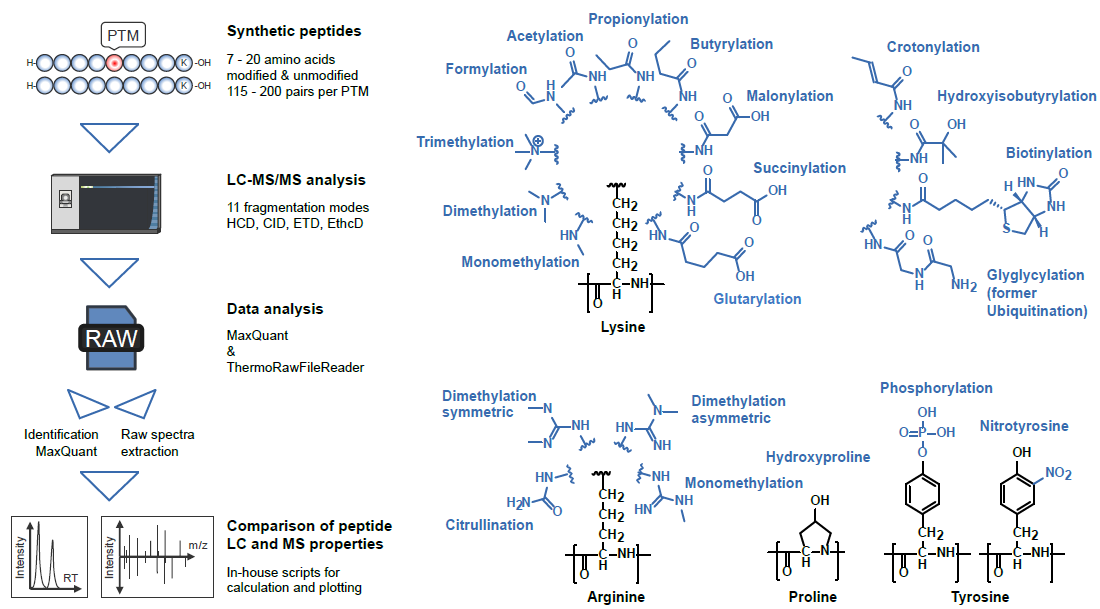
| ProteomeTools: Systematic characterization of 21 post-translational protein modifications by LC-MS/MS using synthetic peptides We are happy to present our newest study on modified peptides: "To more broadly understand the LC-MS/MS characteristics of PTMs, we synthesized and analyzed ~5,000 peptides representing 21 different naturally occurring modifications of lysine, arginine, proline and tyrosine side chains and their unmodified counterparts. The analysis identified changes in retention times, shifts of precursor charge states and differences in search engine scores between modifications. PTM-dependent changes in the fragmentation behavior were evaluated using eleven different fragmentation modes or collision energies. We also systematically investigated the formation of diagnostic ions or neutral losses for all PTMs, confirming 10 known and identifying 5 novel diagnostic ions for lysine modifications." | |
| ProteomeTools @ ASMS 2018 Conference | 04/30/2018 |
| Annual meeting of the American Society for Mass Spectrometry The ProteomeTools team will be present at the ASMS 2018 Conference in San Diego (CA, USA), June 3rd to 7th. We are looking forward to presenting new developments and to discussing the project. Join us for the following presentations: Talks:
Poster:
For abstracts and additional information, please refer to the ASMS 2018 Final Program. | |
| New Research Article | 02/04/2018 |
Mining the human tissue proteome for protein citrullination. We are happy to present a research paper the ProteomeTools project contributed to: " Citrullination is a post-translational modification of arginine catalyzed by five peptidylarginine deiminases (PADs) in humans. The loss of a positive charge may cause structural or functional alterations and while the modification has been linked to several diseases including rheumatoid arthritis and cancer, its physiological or pathophysiological roles remain largely unclear. In part this is owing to limitations in available methodology able to robustly enrich, detect and localize the modification. In this study, we mined data from mass spectrometry-based deep proteomic profiling of 30 human tissues to identify citrullination sites on endogenous proteins. Because citrullination is easily confused with deamidation, we synthesized ~2,200 citrullinated and 1,300 deamidated peptides to build a library of reference spectra. This led to the validation of 375 citrullination sites on 209 human proteins. Further analysis showed that >80% of the identified modifications sites were new and for 56% of the proteins, citrullination was detected for the first time." Read the article at publishers DOI:10.1074/mcp.RA118.000696 | |
| New Research Article | 09/09/2017 |
PROCAL: A set of 40 peptide standards for retention time indexing, column performance monitoring and collision energy calibration. We are happy to present PROCAL, a peptide standard developed withing the ProteomeTools project: "We describe PROCAL (ProteomeTools Calibration Standard), a set of 40 synthetic peptides that span the entire hydrophobicity range of tryptic digests, enabling not only accurate determination of retention time indices but also monitoring of chromatographic separation performance over time. The fragmentation characteristics of the peptides can also be used to calibrate and compare collision energies between mass spectrometers." | |

| ProteomeTools @ HUPO 2017, Dublin, Ireland | 09/01/2017 |
| HUPO 2017 | 16th Human Proteome Organisation World Congress The ProteomeTools team will be present at the 16th Human Proteome Organisation World Congress (HUPO), Sept 17th to 21st. We are looking forwards to discussing the project. Poster ID: 465Abstract Title: Building ProteomeTools based on a complete synthetic human proteome | |
| ProteomeTools ASMS 2017 Conference | 04/25/2017 |
| Annual meeting of the American Society for Mass Spectrometry The ProteomeTools team will be present at the ASMS 2017 Conference in Indianapolis (IN, USA), June 4nd to 8th. We are looking forwards presenting new developments and to discussing the project. | |

| ProteomeTools @ Proteomic Forum 2017 Conference | 03/13/2017 |
| Annual meeting of the German Society for Proteome Research The ProteomeTools team will be present at the Proteomic Forum 2017 Conference in Potsdam (Germany), April 2nd to 5th. We are looking forwards to discussing the project. Poster ID: P050 | |
| News and Views about ProteomeTools | 02/28/2017 |
| Synthetic human proteomes for accelerating protein research The ProteomeTools project and other efforts to cover the human proteome with synthetic peptides standards are featured in a News and Views article in Nature Methods. | |
| ProteomeTools' first paper published in Nature Methods | 01/30/2017 |
Building ProteomeTools based on a complete synthetic human proteome. "We describe ProteomeTools, a project building molecular and digital tools from the human proteome to facilitate biomedical research. Here we report the generation and multimodal liquid chromatography–tandem mass spectrometry analysis of >330,000 synthetic tryptic peptides representing essentially all canonical human gene products, and we exemplify the utility of these data in several applications. The resource (available at www.proteometools.org) will be extended to >1 million peptides, and all data will be shared with the community via ProteomicsDB and ProteomeXchange." | |
